Homodimerization and Annihilation¶
This is for an integrated test of E-Cell4. Here, we test homodimerization and annihilation.
In [1]:
%matplotlib inline
import numpy
from ecell4 import *
from ecell4.extra.ensemble import ensemble_simulations
Parameters are given as follows. D, radius, N_A, and
ka_factor mean a diffusion constant, a radius of molecules, an
initial number of molecules of A, and a ratio between an intrinsic
association rate and collision rate defined as ka andkD below,
respectively. Dimensions of length and time are assumed to be
micro-meter and second.
In [2]:
D = 1
radius = 0.005
N_A = 60
ka_factor = 0.1 # 0.1 is for reaction-limited
In [3]:
N = 30 # a number of samples
rng = GSLRandomNumberGenerator()
rng.seed(0)
Calculating optimal reaction rates. ka is intrinsic, kon is
effective reaction rates. Be careful about the calculation of a
effective rate for homo-dimerization. An intrinsic must be halved in the
formula. This kind of parameter modification is not automatically done.
In [4]:
kD = 4 * numpy.pi * (radius * 2) * (D * 2)
ka = kD * ka_factor
kon = ka * kD / (ka + kD)
Start with A molecules, and simulate 3 seconds.
In [5]:
y0 = {'A': N_A}
duration = 3
T = numpy.linspace(0, duration, 21)
opt_kwargs = {'xlim': (T.min(), T.max()), 'ylim': (0, N_A)}
Make a model with an effective rate. This model is for macroscopic simulation algorithms.
In [6]:
with species_attributes():
A | B | C | {'radius': str(radius), 'D': str(D)}
with reaction_rules():
A + A > ~A2 | kon * 0.5
m = get_model()
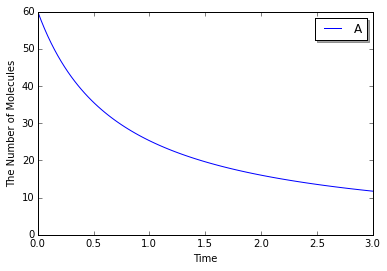
Save a result with ode as obs, and plot it:
In [7]:
obs = run_simulation(numpy.linspace(0, duration, 101), y0, model=ode.ODENetworkModel(m),
return_type='observer', solver='ode')
viz.plot_number_observer(obs, **opt_kwargs)
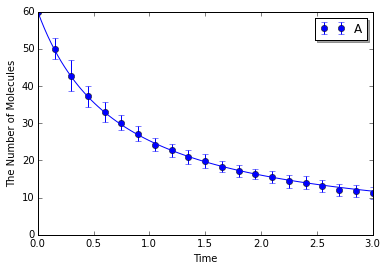
Simulating with gillespie:
In [8]:
ensemble_simulations(T, y0, model=m, return_type='matplotlib',
opt_args=('o', obs, '-'), opt_kwargs=opt_kwargs,
solver='gillespie', n=N)
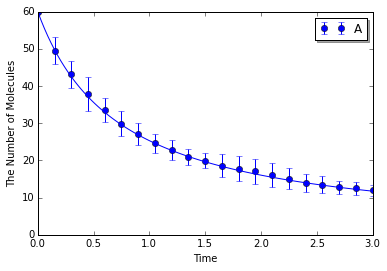
Simulating with meso:
In [9]:
ensemble_simulations(T, y0, model=m, return_type='matplotlib',
opt_args=('o', obs, '-'), opt_kwargs=opt_kwargs,
solver=('meso', Integer3(1, 1, 1), 0.25), n=N)
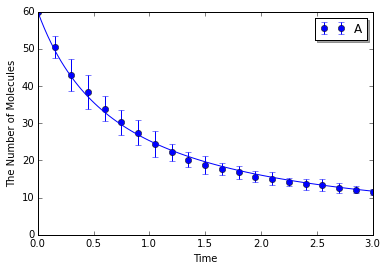
Make a model with an intrinsic rate. This model is for microscopic (particle) simulation algorithms.
In [10]:
with species_attributes():
A | B | C | {'radius': str(radius), 'D': str(D)}
with reaction_rules():
A + A > ~A2 | ka
m = get_model()
Simulating with spatiocyte:
In [11]:
ensemble_simulations(T, y0, model=m, return_type='matplotlib',
opt_args=('o', obs, '-'), opt_kwargs=opt_kwargs,
solver=('spatiocyte', radius), n=N)
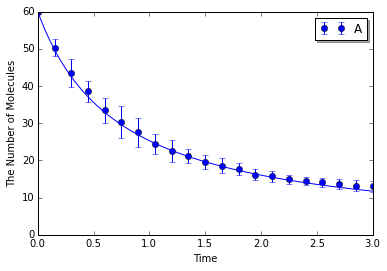
Simulating with egfrd:
In [12]:
ensemble_simulations(T, y0, model=m, return_type='matplotlib',
opt_args=('o', obs, '-'), opt_kwargs=opt_kwargs,
solver=('egfrd', Integer3(4, 4, 4)), n=N)